Research on the unique metabolic mechanisms
Syntrophic propionate oxidation under methanogenic conditions
Post-genomic analytical technique applicable for investigation of syntrophic microorganisms in methane fermentation
Methane fermentation is proceeded by the cooperation of several different characteristics of microorganism. These microorganisms compete and cooperate with each other under an anaerobic environment. We focus on the possibility of acquisition of new functions when the two different microorganisms, propionate-oxidizing bacteria, Pelotomaculum thermopropionicum SI, and hydrogenotrophic methanogen, Methanothermobacter sp. CaT2 constructing a syntrophic association, to reveal the syntrophic mechanisms. In addition, to understand the principle of two different biological systems how to unite or repel and the mechanisms to form a new system. Methane fermenter becomes unstable due to the accumulation of organic acids, especially on short-chain volatile fatty acids. The reason why the degradation of these organic acids requires the syntrophic association of different microorganisms. This association is accomplished based on the molecular functions of these microorganisms.
Propionate oxidation is enhanced by syntrophy
Propionate, one of the volatile fatty acids, is easily accumulated in a methane fermentation due to the unfavorable reaction of its oxidizing endergonic reaction under the standard condition. Hydrogen produced from the reaction need for kept at the low concentration required for cell growth under the condition.
The oxidation reaction of propionic acid, which is one of the volatile fatty acids that easily accumulates in a methane fermentation tank, to acetic acid is an endergonic reaction in the standard condition and is unlikely to proceed spontaneously. This oxidation reaction is performed under low hydrogen partial pressure (or formic acid concentration) that generated from acid oxidation reaction. Otherwise, these microorganisms cannot grow. Therefore, the consumption of hydrogen by hydrogenotrophic methanogens is important. In this way, the coexistence of two different microorganisms is required for to produce methane from propionic acid and acquire energy from each other.

We believe that this syntrophic system produces methane from propionate can be a model for emergent functional expression by the coexistence of two different species. Therefore, we are trying to clarify the metabolism and mechanism of how each microorganism produces methane from propionic acid. Furthermore, we have acquired knowledge on the formation process of aggregates that are considered to play an important role in this symbiosis and the molecules involved in the formation.
Methylmalonyl-CoA pathway is used for anaerobic prpionate oxidation
Anaerobic propionate oxidation in the absence of electron acceptors proceeds via the methylmalonyl CoA pathway (MMC pathway). Propionate is oxidized to acetate via methyl malonyl CoA as an intermediate metabolite, via succinate, fumarate, oxaloacetate, and pyruvate (see the figure below). Such propionate oxidation pathways have been analyzed based on metabolic and genomic analyses of Syntrophobacter fumaroxydance at medium temperature (around 37 °C) bacteria (Plugge et al., 2012) and Pelotomaculum thermopropionicum growing at high temperature (around 55 °C). The expressed proteins were analyzed and the pathway was predicted that this pathway was actually used in P. Thermopropionicum (Kosaka et al., 2006). Furthermore, the whole genome sequence was analyzed and the genetic information of P. Thermopropionicum was revealed (Kosaka et al., 2008). These analyses suggest that syntrophic propionate-oxidizing bacteria have adapted their environment and evolved their genomes.
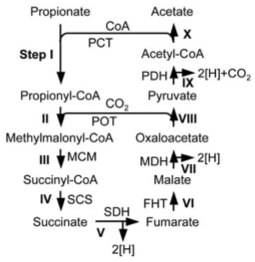
methylmalonyl-CoA pathway
Relationship between microbial symbiosis and aggregation ability, and analysis of the aggregation mechanisms
It has been suggested that different microorganisms aggregate to improve the efficiency of syntrophic propionate-oxidizing metabolism (Ishii et al., 2005). We isolated a thermophilic hydrogenotrophic methanogen Methanothermobacter sp. CaT2 capable of aggregation and reported the genome sequence (Kosaka et al., 2013). This methanogen has surface sugars and forms self-aggregates (Kosaka et al., 2014). The analysis of the cellular factors involved in this aggregation suggested that the protein localized in the surface layer is greatly involved in the aggregation of this methanogenic bacterium by obtaining the mutant strain. It was also suggested that divalent metal ions such as Mg2+ and Ca2+ are involved in the aggregating mechanism (Sumikawa et al., 2019). On the other hand, it was suggested that factors other than proteins are involved in the aggregation enhancement of a mutant strainace (Sumikawa et al., 2020).